Analyzing with OMOP
The Convert service can output data in accordance with the Observational Medical Outcomes Partnership (OMOP) Common Data Model format. The OMOP model standardizes the structure and content of observational data, enabling you to answer the most frequent analytical questions.
Benefits of OMOP
The OMOP conversion is useful for any application that prefers tabular data (e.g., separate, linked data tables) over the nested FHIR JSON/XML format. You can easily ingest the data into a structured database or other off-the-shelf tools, getting you up and running quickly with your own analytics environment. OMOP data is also designed for analytics, presenting data in an easily consumable format with a standard vocabulary for each domain (e.g., all Conditions use SNOMED).
The Convert service uses v5.4 of the OMOP Common Data Model (CDM) specification, with a few exceptions noted in the Convert API. You can alternately request CDM v5.3.
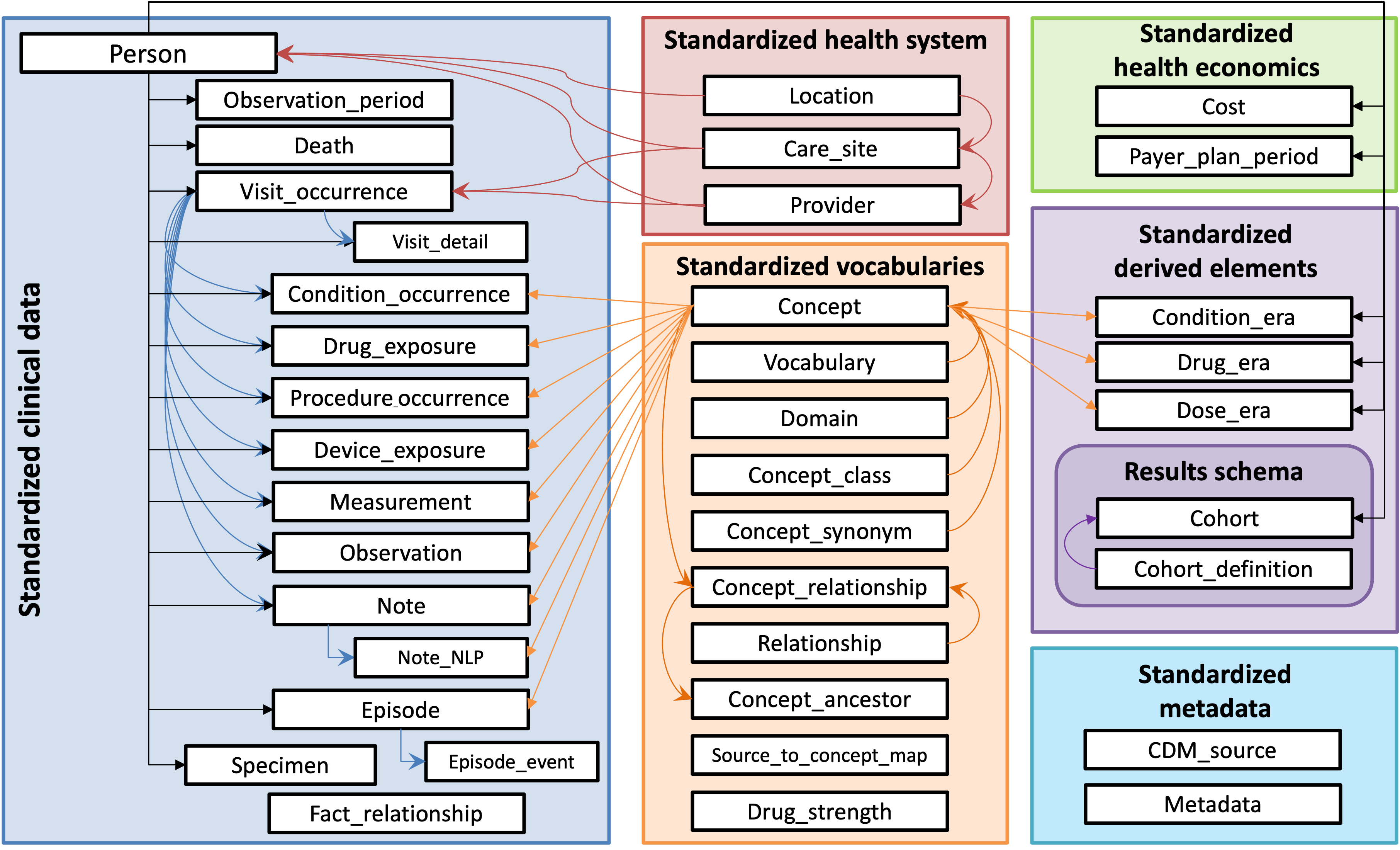 OMOP Common Data Model (source: OHDSI)
OMOP Common Data Model (source: OHDSI)
Converting from FHIR to OMOP
The Convert API will convert a FHIR R4 bundle into OMOP format.
You can use this in conjunction with other convert APIs, allowing data to flow from one conversion to another. For example:
Using OMOP Data
OHDSI has links and tutorials for numerous off-the-shelf software tools that can process OMOP data.
The CSV data can also be readily imported to an SQL database. You can use the scripts provided by OMOP to help you set up the database.
- Find the SQL scripts for your CDM version and database.
- Run the “DDL” script to create the tables.
- Import the converted OMOP output into the tables.
- Run the “primary keys” script to add the primary key constraints to the tables.
- If desired, run the “indices” script to generate SQL indices for more efficient queries.
Note
It is recommended that you not run the “constraints” script. The converted OMOP data will not necessarily comply with all of the additional constraints imposed by this script.
Once the data is imported, you can run standard SQL queries to analyze the data. Some examples are given below.
Data Tables
The output of the OMOP conversion is a ZIP archive containing multiple CSV files, one for each supported OMOP data table.
Tip
See the
OMOP CDM v5.4 specification for details about the data contained within each table. Note that some OMOP-defined tables are omitted from the converted output.
The converted OMOP output includes the following data tables:
Standardized Clinical Data Tables
Standardized Health System Data Tables
Standardized Vocabulary Tables
Custom Tables
Query Examples
Below are some example SQL queries to get you started with common use cases. These examples are from CDM v5.4.
Querying Conditions
Build a list of conditions from the CONDITION_OCCURRENCE table matching diagnosis codes in a value set based on the SNOMED hierarchy. In this example: all descendants of ‘201826 Type 2 diabetes mellitus’.
select * from condition_occurrence co
join concept c on c.concept_id = co.condition_concept_id
join concept_ancestor ca on ca.descendant_concept_id = c.concept_id and ca.ancestor_concept_id = 201826
-- 201826 Type 2 diabetes mellitus
select * from condition_occurrence co
join concept c on c.concept_id = co.condition_concept_id
join concept_ancestor ca on ca.descendant_concept_id = c.concept_id and ca.ancestor_concept_id = 201826
-- 201826 Type 2 diabetes mellitus
Querying Patients
Build a list of patients who have a a CONDITION_OCCURRENCE matching conditions in a value set. In this example: ‘201826 Type 2 diabetes mellitus’.
select p.person_id, c.concept_code, c.concept_name, c.vocabulary_id, count(distinct co.condition_occurrence_id) from person p
join condition_occurrence co on co.person_id = p.person_id
join concept c on c.concept_id = co.condition_concept_id
join concept_ancestor ca on ca.descendant_concept_id = c.concept_id and ca.ancestor_concept_id = 201826
-- 201826 Type 2 diabetes mellitus
group by p.person_id, c.concept_code, c.concept_name, c.vocabulary_id
select p.person_id, c.concept_code, c.concept_name, c.vocabulary_id, count(distinct co.condition_occurrence_id) from person p
join condition_occurrence co on co.person_id = p.person_id
join concept c on c.concept_id = co.condition_concept_id
join concept_ancestor ca on ca.descendant_concept_id = c.concept_id and ca.ancestor_concept_id = 201826
-- 201826 Type 2 diabetes mellitus
group by p.person_id, c.concept_code, c.concept_name, c.vocabulary_id
Querying Labs
Build a list of lab measurements (from the MEASUREMENT table) by LOINC component (e.g., A1C tests).
select * from measurement m
join concept c on m.measurement_concept_id = c.concept_id
join concept_relationship cr on c.concept_id = cr.concept_id_1
join concept vsc on vsc.concept_id = cr.concept_id_2 and cr.relationship_id = 'Has component'
and vsc.concept_class_id = 'LOINC Component' and vsc.concept_name in ('Hemoglobin A1c/Hemoglobin.total')
select * from measurement m
join concept c on m.measurement_concept_id = c.concept_id
join concept_relationship cr on c.concept_id = cr.concept_id_1
join concept vsc on vsc.concept_id = cr.concept_id_2 and cr.relationship_id = 'Has component'
and vsc.concept_class_id = 'LOINC Component' and vsc.concept_name in ('Hemoglobin A1c/Hemoglobin.total')
Querying Medications
Build a list of medications organized by frequency of use (per ingredient) based on the DRUG_EXPOSURE table.
select vsc.concept_name,count(distinct d.person_id) as countPersons, count(distinct d.drug_exposure_id) as countMeds from drug_exposure d
join concept_relationship cr on cr.concept_id_2 = d.drug_concept_id and cr.relationship_id = 'RxNorm ing of'
join concept vsc on cr.concept_id_1 = vsc.concept_id and vsc.concept_class_id = 'Ingredient'
group by vsc.concept_name
order by countPersons desc
select vsc.concept_name,count(distinct d.person_id) as countPersons, count(distinct d.drug_exposure_id) as countMeds from drug_exposure d
join concept_relationship cr on cr.concept_id_2 = d.drug_concept_id and cr.relationship_id = 'RxNorm ing of'
join concept vsc on cr.concept_id_1 = vsc.concept_id and vsc.concept_class_id = 'Ingredient'
group by vsc.concept_name
order by countPersons desc
Retrieve Value Set Members
There are several ways to query for the members of a value set. This example filters by LOINC component via the CONCEPT_RELATIONSHIP table.
select * from concept vsc
join concept_relationship cr on cr.concept_id_1 = vsc.concept_id and cr.relationship_id = 'Component of'
join concept c on cr.concept_id_2 = c.concept_id
where vsc.concept_name = 'Hemoglobin A1c/Hemoglobin.total' and vsc.vocabulary_id = 'LOINC' and vsc.concept_class_id = 'LOINC Component'
--vsc.concept_name = 'Hemoglobin A1c/Hemoglobin.total'
--vsc.concept_name = 'SARS-CoV-2 (COVID-19) Ag'
select * from concept vsc
join concept_relationship cr on cr.concept_id_1 = vsc.concept_id and cr.relationship_id = 'Component of'
join concept c on cr.concept_id_2 = c.concept_id
where vsc.concept_name = 'Hemoglobin A1c/Hemoglobin.total' and vsc.vocabulary_id = 'LOINC' and vsc.concept_class_id = 'LOINC Component'
--vsc.concept_name = 'Hemoglobin A1c/Hemoglobin.total'
--vsc.concept_name = 'SARS-CoV-2 (COVID-19) Ag'
This example filters by RxNorm Ingredient via the CONCEPT_RELATIONSHIP table.
select * from concept vsc
join concept_relationship cr on cr.concept_id_1 = vsc.concept_id and cr.relationship_id = 'RxNorm ing of'
join concept c on cr.concept_id_2 = c.concept_id
where vsc.concept_class_id = 'Ingredient' and vsc.concept_name ilike '%caduet%'
select * from concept vsc
join concept_relationship cr on cr.concept_id_1 = vsc.concept_id and cr.relationship_id = 'RxNorm ing of'
join concept c on cr.concept_id_2 = c.concept_id
where vsc.concept_class_id = 'Ingredient' and vsc.concept_name ilike '%caduet%'
This example uses SNOMED hierarchical relationships. However, note that it is not recursive and only returns immediate children.
select * from concept c
join concept_relationship cr on cr.concept_id_1 = 201826 and cr.relationship_id = 'Subsumes'
-- 201826 Type 2 diabetes mellitus
select * from concept c
join concept_relationship cr on cr.concept_id_1 = 201826 and cr.relationship_id = 'Subsumes'
-- 201826 Type 2 diabetes mellitus
OMOP vocabulary tables also provide a way to get all descendants within the SNOMED hierarchy using the CONCEPT_ANCESTOR table. You are even able to specify the number of generations to include if desired.
select * from concept c
join concept_ancestor ca on ca.descendant_concept_id = c.concept_id and ca.ancestor_concept_id = 201826
-- 201826 Type 2 diabetes mellitus
select * from concept c
join concept_ancestor ca on ca.descendant_concept_id = c.concept_id and ca.ancestor_concept_id = 201826
-- 201826 Type 2 diabetes mellitus